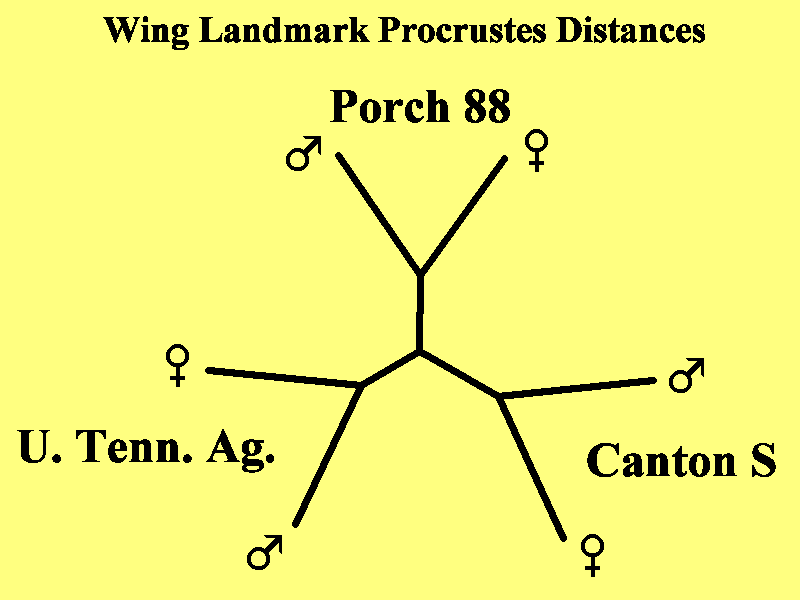

Fig. 5. Unrooted phenetic similarity tree of wing landmark
sexual dimorphism of three wildtype Drosophila
melanogaster strains. The 'best' tree of morphological distances
was computed based on the Kitch algorithm of Felsenstein
(1989) applied to pair wise Procrustes distances computed using
the tpsSmall algorithm applied to the
set of six average landmark sets computed for the two sexes of three
wild type D. melanogaster strains. A
(default) Kitch analysis was performed on the Procrustes difference
matrix (Table I) using the Fitch-Margoliash
method with contemporary tips. Negative branch lengths were
not allowed and 123 trees were examined. The
Drawtree algorithm was used to create the unrooted tree using computed
limb lengths. Subsequent analysis of 300
wings of the three stains concluded that there is no significant
difference in the observed sexual dimorphism
between the three stains.
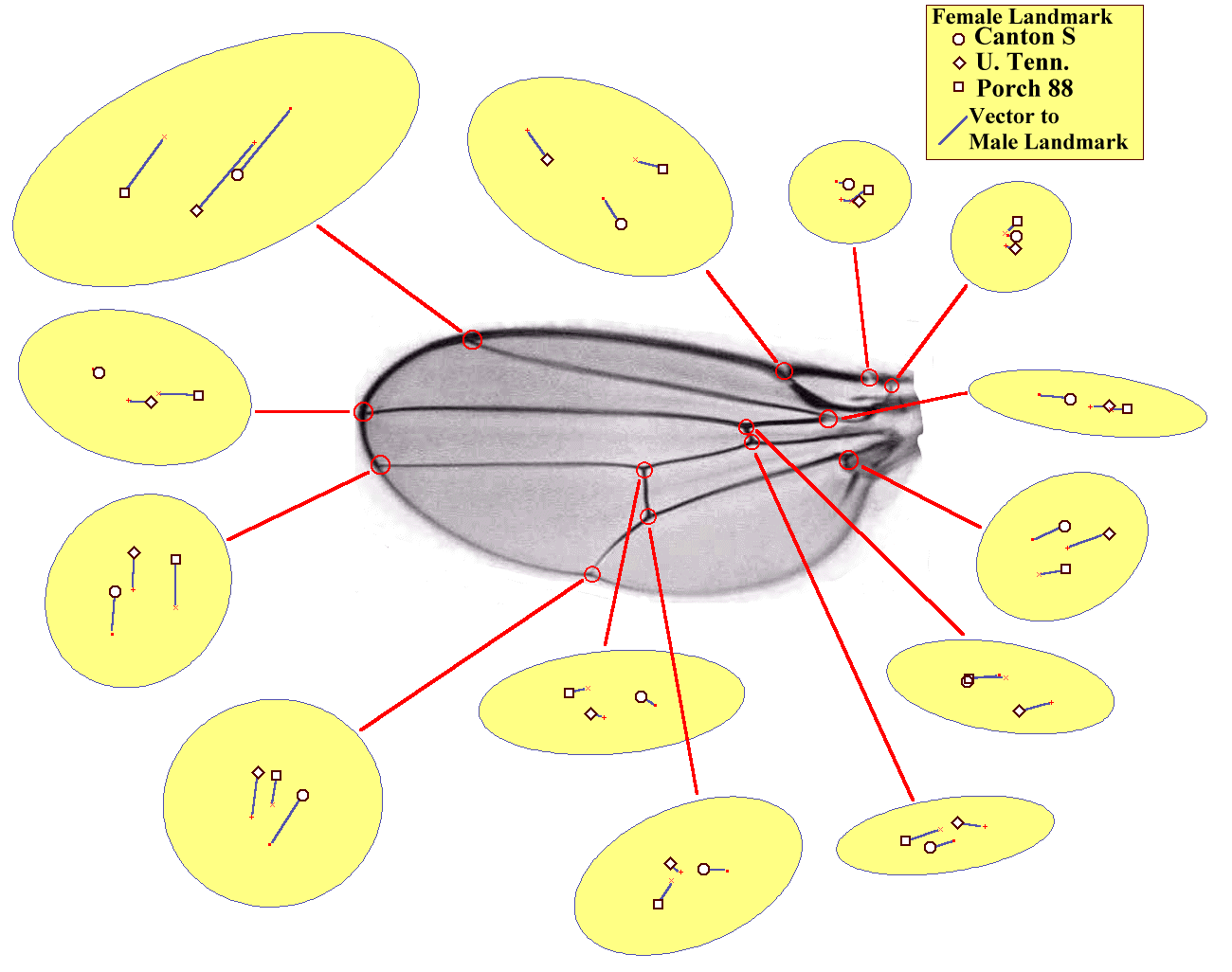
Figure 6. Female landmarks and vector difference to male landmarks for 13 wingvein landmarks in three Wild Type strains of Drosophila melanogaster. Inset ellipses are approximate 95% confidence intervals for the raw aligned landmarks. The female averages (hollow square, circle and diamond) and male averages (solid square, plus and cross) are averages of landmarks, aligned using the least square algorithm of Siegel (1983) (view the image alone for 2X resolution.)